Studying protein interactions of spike glycoprotein and ACE2 receptor
Understanding the spike-ACE2 receptor binding
The outbreak of the SARS-CoV-2 pandemic has prompted extensive research into understanding how the virus enters host cells.
The viral infection involves the transmembrane spike glycoprotein, which forms homotrimers on the viral surface. These spike proteins interact tightly with the human angiotensin-converting enzyme 2 (ACE2), acting as a functional receptor for the virus. The spike-ACE2 interaction initiates a series of conformational changes that enable the virus to fuse with the host cell membrane.
How to study the oligomeric state of the spike glycoprotein
Mass photometry is a powerful bioanalytical technique that allows scientists to measure the mass of biomolecules in solution. It has proven to be particularly valuable in studying the oligomeric states of proteins and quantifying protein-protein interactions. In the context of the SARS-CoV-2 proteins, mass photometry was used to study the spike protein and its interaction with the ACE2 receptor, which is believed to be the primary route of viral entry into human cells.
Anti-spike antibody interactions with spike glycoprotein
As mass photometry enables study of proteins in 30 kDa – 5 MDa range, it can also be used to characterise antibody-spike protein interaction or screen anti-spike antibodies for their affinity/potency. This knowledge could be instrumental in the development of therapeutic interventions aimed at preventing viral entry into host cells.
To learn how mass photometry can be used to study protein oligomerization and protein-protein interactions
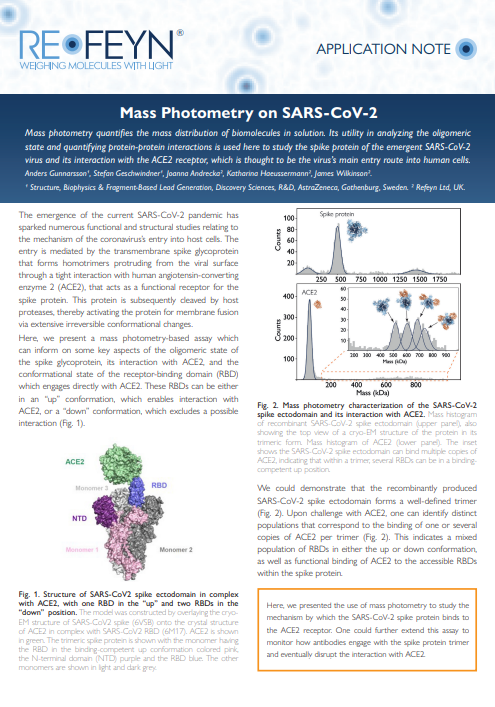
Mass photometry on SARS-CoV-2
Additional resources
WEBINAR: Probing Antibody Binding to the SARS-CoV-2 Spike Protein
In this webinar, Dr. Victor Yin, a postdoctoral researcher of biomolecular mass spectrometry and proteomics at Utrecht University, will discuss how mass photometry and charge detection mass spectrometry enable the study of several challenging protein systems, including the interaction of full antibodies with SARS-CoV-2.

BLOG POST: SARS-CoV-2 studies highlight mass photometry’s ability to characterise complex heterogeneous systems
Mass photometry has proved valuable in coronavirus research, especially in characterising complex and heterogeneous proteins, and in analysing protein interactions and stoichiometry. In this blog post, we highlight ways that mass photometry has helped investigate previously intractable questions and advance our understanding of SARS-CoV-2.
To discuss how mass photometry can facilitate your viral protein research
More Application Notes
Browse through our catalogue of application notes highlighting some recent case studies featuring mass photometry.
