How to characterize protein oligomerization with mass photometry
Role of the oligomeric state of a protein
Many proteins self-assemble into quaternary structures formed by several subunits, a process known as oligomerization. Different oligomeric states of a protein can modulate its functionality, such as enzymatic activity, influence gene regulation or functionality of some membrane proteins. Finally, altered oligomeric states can lead to protein disfunction, which links with the development of some diseases.
Therefore, developing techniques that characterize the oligomeric state of proteins and efficiently determine the factors that define protein oligomerization is important for both applied and translational research. However, characterizing protein oligomeric states is not always straightforward, especially in cases where some oligomeric species are present in very low concentrations.

Automated mass photometry supports protein oligomerization studies
Mass photometry is a single-molecule technique that uses light to measure the mass of every molecule that lands on a glass measuring surface. Thanks to its high resolution and single-molecule approach, mass photometry gives the complete mass distribution of the sample within the working range of the instrument, including rare species that form less than 1% of the main sample population.
Mass photometry makes it easy to test the factors that define the oligomerization behavior of proteins. It requires small volumes of sample at low concentrations, and measurements take only minutes from sample loading to results. Sample preparation is straightforward, and mass photometry is compatible with a wide range of commonly used buffers.
Further supporting protein oligomerization studies, automated platforms are available for mass photometers. The TwoMP Auto can measure up to 24 samples autonomously, with higher consistency than a human operator. This makes automated mass photometry ideal for studies that explore how a given factor defines oligomerization behavior – such as the concentration of the target protein itself or that of an effector molecule.
To find out how mass photometry streamlines oligomerization studies
Characterizing protein oligomerization with automated mass photometry
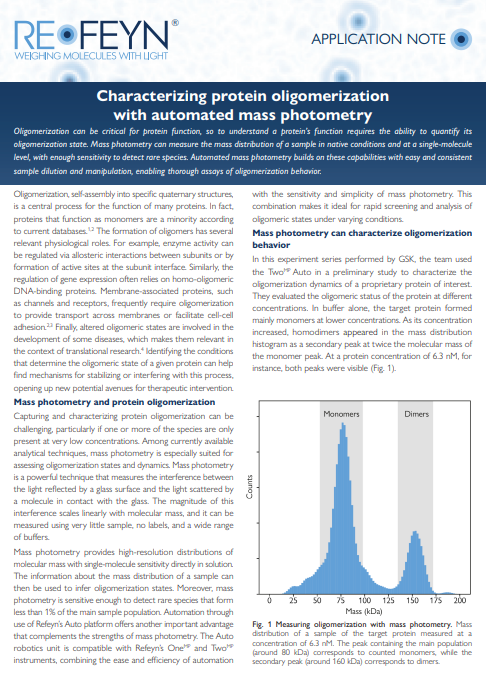
Learn about the advantages of automated mass photometry for oligomerization studies. Read the application note and discover how the quick and repeatable measurements provided by the TwoMP Auto help characterize the factors that define oligomerization behavior. Look at the data from two case studies in which protein concentration or the presence of an effector is varied, and the treated samples are systematically characterized with automated mass photometry. Here, you will see how the single-molecule mass distributions provided by mass photometry can be used to identify the different oligomeric species in a sample.
Additional resources
BLOG: Blog: Studying protein oligomerization with mass photometry – insights from a future PI
In this blog post, we talk to Dr. Kathryn Gunn about her research into the oligomeric states of lipoprotein lipase and how its structure relates to its activity. Her team used mass photometry to analyze the LPL oligomeric states and structural changes in solution, while cryoEM was used to visualize LPL structures in thin ice layers. Cross-linking, mass spectrometry, and western blotting were also employed to further characterize LPL oligomers and protein interactions.


WEBINAR: Automated mass photometry: Easing the path to biomolecular characterization
In this webinar, we discuss in detail the automated mass photometry and show data on some of its most attractive applications—such as screening and titration assays. We illustrate how automation and the associated improvement in reproducibility can make biomolecular characterization – including protein oligomerization – with mass photometry even easier.
To learn more about how mass photometry empowers your oligomerization studies
More Application Notes
Browse through our catalogue of application notes highlighting some recent case studies featuring mass photometry.
