Monitoring protein folding with mass photometry
Monitoring protein folding and unfolding
Chemical denaturation is a process that disrupts the native structure of proteins, using denaturants like urea and guanidine hydrochloride. These denaturants promote protein unfolding by exposing hydrophobic residues to the surrounding aqueous solution.
Mass photometry is a single-molecule method that measures the mass of biomolecules in solution, and is insensitive to changes in the secondary structure of proteins. Whether a protein is denatured or native, its mass remains constant – however its behaviour might change
How to monitor protein refolding dynamics?
Mass photometry is a valuable tool for monitoring the refolding dynamics of proteins, particularly in the context of denaturing conditions. When denaturants disrupt protein structures, mass photometry can capture changes in the behavior of molecules by measuring biomolecular mass. More specifically, you can observe whether a denaturant has disrupted complex formation or the oligomerization behavior of a protein of interest.
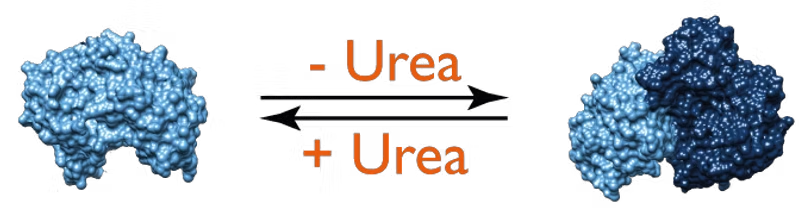
Image: The presence of urea disrupts enolase dimerization. For experimental details, download the app note
A case study monitoring protein renaturation of an enzyme, enolase, shows the value of mass photometry. In native conditions, enolase presents itself mostly in a dimeric form. Mass photometry showed that exposure to denaturants disrupts protein dimerization. It also allowed monitoring of renaturation of the enolase samples – with decreasing concentrations of denaturants, enolase resumed forming dimers.
Mass photometry can therefore provide insights into protein-protein interactions and how they are affected by denaturants or other stressors, making mass photometry a valuable tool for studying protein structure and behavior.
To learn how mass photometry can measure protein denaturation and renaturation
Mass photometry in denaturing conditions
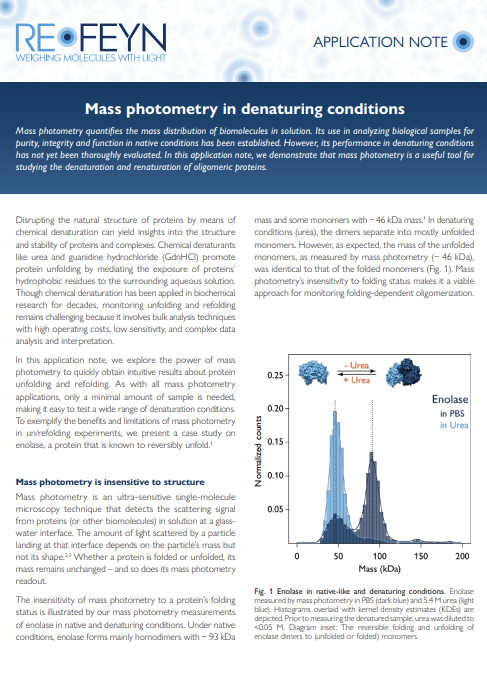
Learn how mass photometry can monitor protein stability. Discover how it can provide valuable insights into protein unfolding and refolding processes, even when traditional bulk methods fall short. Explore a case study involving the reversible unfolding of enolase, to understand how mass photometry excels in monitoring the denaturation and renaturation of proteins within multimeric complexes. Learn also what is the impact of denaturant concentrations on optical measurements and the strategies to overcome challenges.
Additional resources
APPLICATION NOTE: Quantifying protein binding affinities using mass photometry
Read this application note to learn how you can use the TwoMP mass photometer to characterize protein-protein interactions, quickly and easily. Discover how you can determine the relative abundance of each protein and the complexes they form in solution, using this label-free bioanalytical tool. Learn also how you can use the mass photometry measurements to calculate the equilibrium dissociation constant (KD) for each interaction.

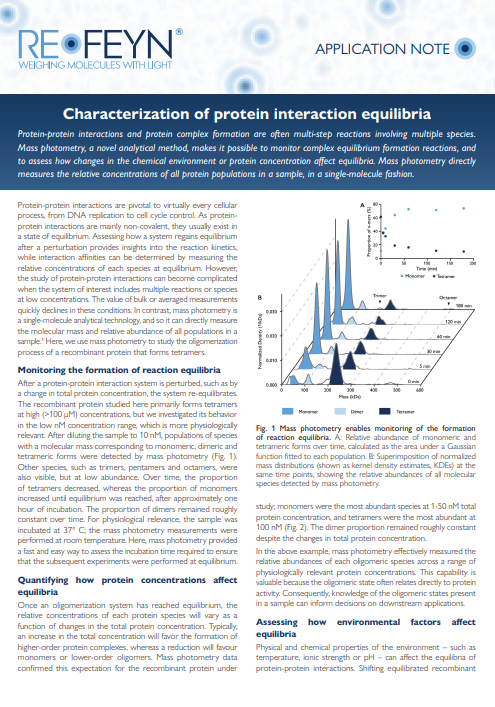
APPLICATION NOTE: Characterization of protein interaction equilibria
Learn how mass photometry can be applied to the study of complex equilibrium formation reactions and assess how changes in the chemical environment or protein concentration affect equilibria. More specifically, discover how mass photometry can be used to study the oligomerization process of a recombinant protein that forms tetramers. Using this multimeric protein as an example, learn how to measure the relative abundances of each oligomeric species across a range of physiologically relevant protein concentrations and assess the effect of environmental factors (such as temperature) on reaction equilibria.
To discuss how mass photometry can be applied to the measurement of protein unfolding and refolding
More Application Notes
Browse through our catalogue of application notes highlighting some recent case studies featuring mass photometry.
